Abstract
In this paper, we present the second release of the GyDB Collection: an in-progress repository of manually refined multiple alignments, Hidden Markov Model (HMM) profiles and Majority-rule consensus (MRC) sequences of viruses and mobile genetic elements. Alignments are available in multiple formats plus a color-shaded HTML for preserved motif visualization. The HTML format includes links to each sequence?s GenBank accession at NCBI. HMM profiles and MRCs are based on each of the accepted protein domain consensus accepted per monophyletic group of mobile genetic elements (MGEs) and protein domain. The collection is the repository of the Gypsy Database (GyDB) of Mobile Genetic Elements and contemplates all protein domains encoded by all phylogenetic subsets of MGEs classified at GyDB and, as such, is subjected to continuous growth.
Availability
Available online July 30, 2009. The GyDB Collection is distributed at URL 1 under the terms of the Creative Commons Atribution license (URL 2), which allows unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Since the existence of mobile DNA was first suggested by McClintock (1) MGEs have been an important object of study in multiple areas of research (2). Nearly all MGEs fit into three major categories of transposons classified by their mechanisms. Class I are retroelements (3) that mediate their transposition life cycle through an RNA-DNA reverse transcription process. This class includes all reverse transcriptase (RT) dependent retrotransposons and retroviruses. Class II are DNA-based transposons that move directly from one position to another in host genomes (4, 5, 6). Class III are the Miniature Inverted-Repeats Transposable Elements (1, 72, 8). Viruses are ubiquitous genetic entities circulating in living organisms and exist as free particles called virions, which are typically RNA or DNA genomes surrounded by a protein shell called a capsid. In the post-genomic era, sequencing projects have allowed the characterization of multiple distinct sequences of viruses and MGEs from different biological organisms. It is now known that many types of viruses evolve from MGEs but viruses are particularly variable in morphology, replication mechanisms, genome organization and many other features. Such diversity derives from the multiple distinct virus origins, which in most cases are yet to be understood. In this regard, sequencing projects have an impressive potential to shed light on biologically relevant questions in viral and MGE evolution. We think that understanding the diversity and the genomic similarities of the multiple distinct viral and MGE systems will lead to new insights in molecular evolution and epidemiology. With this aim, and in an attempt to enhance collaborative knowledge in the field, we built the GyDB project (4), which is a long-term research project for analyzing and classifying non-redundant MGEs based on their evolutionary profiles. The GyDB is accessible at URL 3. In this paper we introduce the GyDB collection: the repository of all multiple alignments, HMM profiles and MRC sequences for viruses and MGEs derived from the GyDB project.
Overview
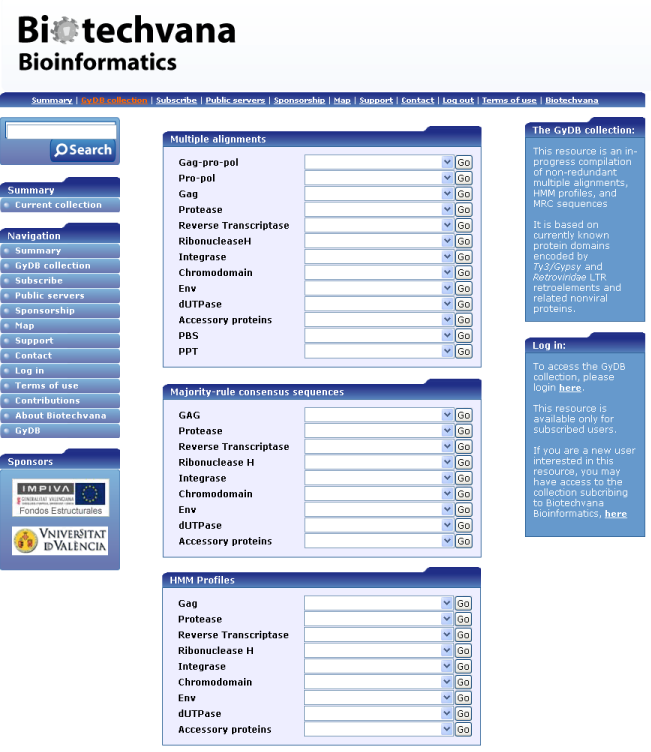
Sequence data analyzed in the GyDB project are usually obtained from the GenBank at the National Center of Biotechnology Information URL 4 or from any other available scientific source. Based on all this material we have built an in-progress collection of multiple alignments and molecular profiles useful in retroelement taxonomy and identification of new MGE species. The collection is presented via a web server divided in three categories: multiple alignments, HMM profiles and MRC sequences. As shown in Figure 1, each category has a set of drop-down lists that display items for multiple alignments, HMM profiles, and MRC sequences grouped by domains.
Multiple Alignments
Multiple alignments are obtained and manually refined using CLUSTAL X 9 and GENEDOC URL 5 based on DNA sequences and protein domains (according to the phylogenetic models introduced in section ?Phylogenies? at GyDB, URL 6). The collection also includes compound alignments based on the concatenation of many (or all) protein domains typically coded by one or more MGE lineages (for instance, all pol polyprotein domains joined together into a single alignment).
In the process of refinement, we use the following groups of amino acid similarity; [T,S small nucleophile amino acids] [K,R,H basic amino acids], [D,E,N,Q acidic amino acid and relative amides], and [L,I,V,M,A,G,P,F,Y,W hydrophobic amino acids]. These groups of similarity take into primary consideration the physiochemical properties of amino acids.
Compound alignments are created using a PHP script (Joint Alignments Server) available under the section ?Scripts? in Biotechvana Bioinformatics (URL 7). Alignments are available in six formats: FASTA, PIR, MSF, Stockholm, Clustal, Phylip plus a color-shaded HTML in order to easily view preserved motifs. The HTML format includes hyperlinks to each sequence GenBank accession at NCBI.
Hidden Markov Model Profiles
Detection of well-defined protein motifs or domains is useful to classify proteins into families and these classifications can be used to assign putative physiological roles to new discovered proteins (10). One of the most powerful methodologies in this area are HMM profiles (11), which are statistical models constructed from multiple sequence alignments. HMM profiles capture position-specific information on the degree of conservation of residues in each column of the alignment. Taking into account the monophyletic clusters reported by GyDB phylogenies, we continuously construct and update the collection of HMM profiles, using HMMER 2.3.2 (URL 8).
Majority-Rule Consensus Sequences
They facilitate identification of relationships and taxonomy of sequences, as well as discernment of conserved motifs that may be characteristic of protein domains. The MRC sequence methodology consists in the creation of a single consensus from a set of aligned sequences. In MRC sequences, highly conserved residues (probability greater than or equal to 90 percent for DNA and greater than or equal to 50 percent for proteins) are shown in uppercase, and less conserved residues in lowercase. We used our collection of generated HMM profiles to construct a derived collection of MRC sequences using HMMER.They facilitate identification of relationships and taxonomy of sequences, as well as discernment of conserved motifs that may be characteristic of protein domains. The MRC sequence methodology consists in the creation of a single consensus from a set of aligned sequences. In MRC sequences, highly conserved residues (probability greater than or equal to 90 percent for DNA and greater than or equal to 50 percent for proteins) are shown in uppercase, and less conserved residues in lowercase. We used our collection of generated HMM profiles to construct a derived collection of MRC sequences using HMMER.
Concluding Remarks
The GyDB Collection contemplates, by monophyletic groups, protein products encoded by viruses and MGEs and the distinct groups of nonviral gene or protein families related to them. Due to high divergence, many of the alignments we provide are manually built. The collection is a work-in-progress attached to the GyDB project. Taking into primary consideration that evidences of new viral and sequences grow parallel to sequencing projects, the GyDB Collection requires a continuous update effort. This is, however, the most interesting aspect of this approach, which is a significant platform to compare and evaluate any new finding in the topic. Reversely, this feedback is useful to us in order to re-build the collection and obtain more accurate profile models of viruses and MGEs.
Acknowledgments
We are grateful to Fernando Gonz?lez, Rosario Gil and Pascual Asensi for technical support and GyDB hosting at ICBIBE of University of Valencia. The research has been partly supported by grant 17092008 from ENISA (Empresa Nacional de Innovacion S.A), and by grants IMCBTA/2005/45, IMIDTD/2006/158, IMIDTD/2007/33, and IMIDTD/2008/103 from IMPIVA.
Literature
- 1. McClintock,B. (1948) Carnegie Inst. Wash. Year book, 47, 155-169.
- 2. Kazazian,H.H., Jr. (2004) Science, 303, 1626-1632.
- 3. Temin,H.M. (1989) Nature, 339, 254-255.
- 4. Llorens,C., Futami,R., Bezemer,D. and Moya,A. (2008) Nucleic Acids Research (NAR), 36 (Database-Issue): 38-46.
- 5. Mizuuchi,K. (1992) Annu. Rev. Biochem., 61, 1011-1051.
- 6. Lerat,E. and Capy,P. (1999) Mol. Biol. Evol., 16, 1198-1207.
- 7. Wessler,S.R., Bureau,T.E. and White,S.E. (1995) Curr. Opin. Genet. Dev., 5, 814-821.
- 8. Bureau,T.E., Ronald,P.C. and Wessler,S.R. (1996) Proc. Natl. Acad. Sci. U.S.A., 93, 8524-8529.
- 9. Thompson,J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) Nucleic Acids Research (NAR), 25, 4876-4882.
- 10. Schug,J., Diskin,S., Mazzarelli,J., Brunk,B.P. and Stoeckert,C.J., Jr. (2002) Genome Res., 12, 648-655.
- 11. Eddy,S.R. (1998) Bioinformatics, 14, 755-763.
- 1. The GyDB Collection: http://gydb.uv.es/biotechvana/bioinformatics/main.php?document=collection
- 2. Creative Commons Attribution license: http://creativecommons.org/licenses/by/2.0
- 3. GyDB: http://gydb.uv.es
- 4. NCBI: http://www.ncbi.nlm.nih.gov
- 5. GENEDOC: http://www.psc.edu/biomed/genedoc
- 6. PR tree: http://gydb.uv.es/gydb/phylogeny.php?tree=pr
- 7. Biotechvana Bioinformatics: http://gydb.uv.es/biotechvana/bioinformatics
- 8. HMMER: http://hmmer.janelia.org